Time-saving method for inventing drugs results in possible treatmentBy Steven Schultz Princeton NJ -- Engineers at the University have developed an accelerated method for inventing protein-based drugs and, in a unique collaboration with two other institutions, have used it to design improved versions of a possible treatment for inflammatory disorders.
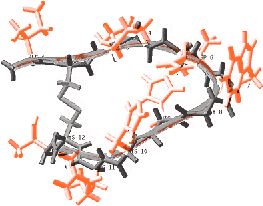
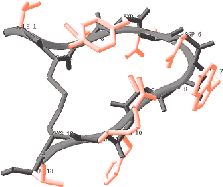
The new approach allows scientists to bypass time-consuming, trial-and-error procedures for making and testing billions of protein fragments as possible drug candidates. Instead, the researchers invented mathematical techniques and used a cluster of computers to calculate the near-optimum compositions for the proteins. The achievement grew out of a long-time collaboration between Christodoulos Floudas, a Princeton professor of chemical engineering, John Lambris, a professor of pathology and laboratory medicine at the University of Pennsylvania, and Dimitrios Morikis, a researcher in chemical and environmental engineering at the University of California-Riverside. The researchers reported their findings in the July 16 issue of the Journal of the American Chemical Society. The project began in 1996 when Lambris and colleagues invented a potential anti-inflammatory drug called compstatin and wanted to adjust its structure to make it more powerful. Compstatin, which has been tested in animals but not humans, inhibits an immune system component called complement, which plays an important role in fighting infections. In some medical conditions, the complement system goes awry and causes damage to the body's own tissue. Complement-related injury, for example, is a major factor in heart attacks, stroke, transplant surgeries, burn injuries and autoimmune diseases. A drug such as compstatin could be useful in treating more than 25 conditions, the researchers said.
Researchers in Lambris' lab sought improved versions of compstatin by creating and testing billions of alternative versions. Compstatin, like other proteins, is a chain of amino acids strung end-to-end. Substituting different amino acids at various points along the chain was a hit-or-miss process. In the meantime, Morikis used nuclear magnetic resonance spectroscopy to determine the three-dimen- sional structure of compstatin, allowing Lambris a first glimpse of his drug's physical appearance. Lambris and Morikis used this information to design several new versions with specific alterations that they predicted would make the drug work better. Combined with Lambris' work making billions of random substitutions, these efforts produced a few slightly improved compstatin analogs. The job of finding dramatic improvements fell to Floudas, whose expertise is in understanding the relation between a protein's amino acid sequence and its three-dimensional structure. When cells make proteins, the chain immediately folds into a complex shape, like a mangled clock spring. It is very hard to predict what shape will result from any particular amino acid sequence. Floudas' challenge was that there are so many possible sequences. Compstatin is a relatively short protein, called a peptide, with only 13 amino acids. However, the body has a repertoire of 20 different amino acids, so substituting every possible amino acid in all 13 slots results in 80 quadrillion possible sequences. "It would have taken us years to synthesize and screen the 80 quadrillion possible peptide sequences that [Floudas'] protein design program considered," said Lambris. To solve the problem, Floudas and postdoctoral associate John Klepeis drew on mixed-integer optimization theory, a branch of mathematics used for finding the best possible value from many variables. Their two-step approach first narrowed the many possibilities to a short list of promising sequences. They then used a technique they had developed earlier, called Astro-Fold, which predicts the shape that will arise from any given sequence. They applied Astro-Fold to their short list and checked to see how closely each sequence resembled compstatin. In the end, Floudas provided 14 possible sequences to Lambris, who made and tested them. Of the 14, eight were three to seven times more effective than the best previous version of compstatin. "This is the first time the system has been used, so it is very pleasant for us to see such results," said Floudas. He also was intrigued to find that the method produced surprises. In the sequences that turned out to be most effective, the program suggested an amino acid substitution that had not been considered in the billions of previous tests. "It really has been a very nice collaboration between theorists and experimentalists," said Floudas. "We now have an excellent basis from which to develop variants with novel properties that have potential to be clinically relevant," said Lambris. "Ultimately, the value of our efforts will be demonstrated with further biological analysis and in the clinic. This collaboration enables us to get there sooner." The researchers believe that the method will be widely applicable to other protein drugs and are planning to continue their collaboration. Princeton and Penn have filed for a joint patent on the new versions of compstatin. Princeton also has filed for a patent on Floudas' sequence discovery method. |
Contents |
|||||||||